Published: 30 March 2023
Multiple intermolecular interactions facilitate rapid evolution of essential genes
作者 | Huei-Yi Lai, Yen-Hsin Yu, Yu-Ting Jhou, Chia-Wei Liao, Jun-Yi Leu*
通讯作者地址 | Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan

Abstract
Essential genes are commonly assumed to function in basic cellular processes and to change slowly. However, it remains unclear whether all essential genes are similarly conserved or if their evolutionary rates can be accelerated by specific factors. To address these questions, we replaced 86 essential genes of Saccharomyces cerevisiae with orthologues from four other species that diverged from S. cerevisiae about 50, 100, 270 and 420 Myr ago. We identify a group of fast-evolving genes that often encode subunits of large protein complexes, including anaphase-promoting complex/cyclosome (APC/C). Incompatibility of fast-evolving genes is rescued by simultaneously replacing interacting components, suggesting it is caused by protein co-evolution. Detailed investigation of APC/C further revealed that co-evolution involves not only primary interacting proteins but also secondary ones, suggesting the evolutionary impact of epistasis. Multiple intermolecular interactions in protein complexes may provide a microenvironment facilitating rapid evolution of their subunits.
必需基因通常被认为在基本的细胞过程中发挥作用,并且变化缓慢。然而,目前尚不清楚是否所有必需基因都具有相似的保守性,或者它们的进化速度是否可以被特定的因素加速。为了解决这些问题,我们将酿酒酵母(Saccharomyces cerevisiae)的86个必需基因替换为约50、100、270和420 Myr前从酿酒酵母(S. cerevisiae)分化而来的其他4个物种的同源基因。我们发现了一组快速进化的基因,它们通常编码大蛋白复合体的亚单位,包括后期促进复合体/环体(APC/C)。通过同时替换相互作用的组分,快速进化基因的不相容性得以挽救,这表明它是由蛋白质共同进化引起的。对APC/C的详细研究进一步揭示了协同进化不仅涉及初级相互作用蛋白,还涉及次级相互作用蛋白,表明上位作用对进化的影响。蛋白质复合物中的多种分子间相互作用可能提供了促进其亚单位快速进化的微环境。
Fig. 1: Functional assays reveal incompatibility between essential gene orthologues and theS. cerevisiae background.
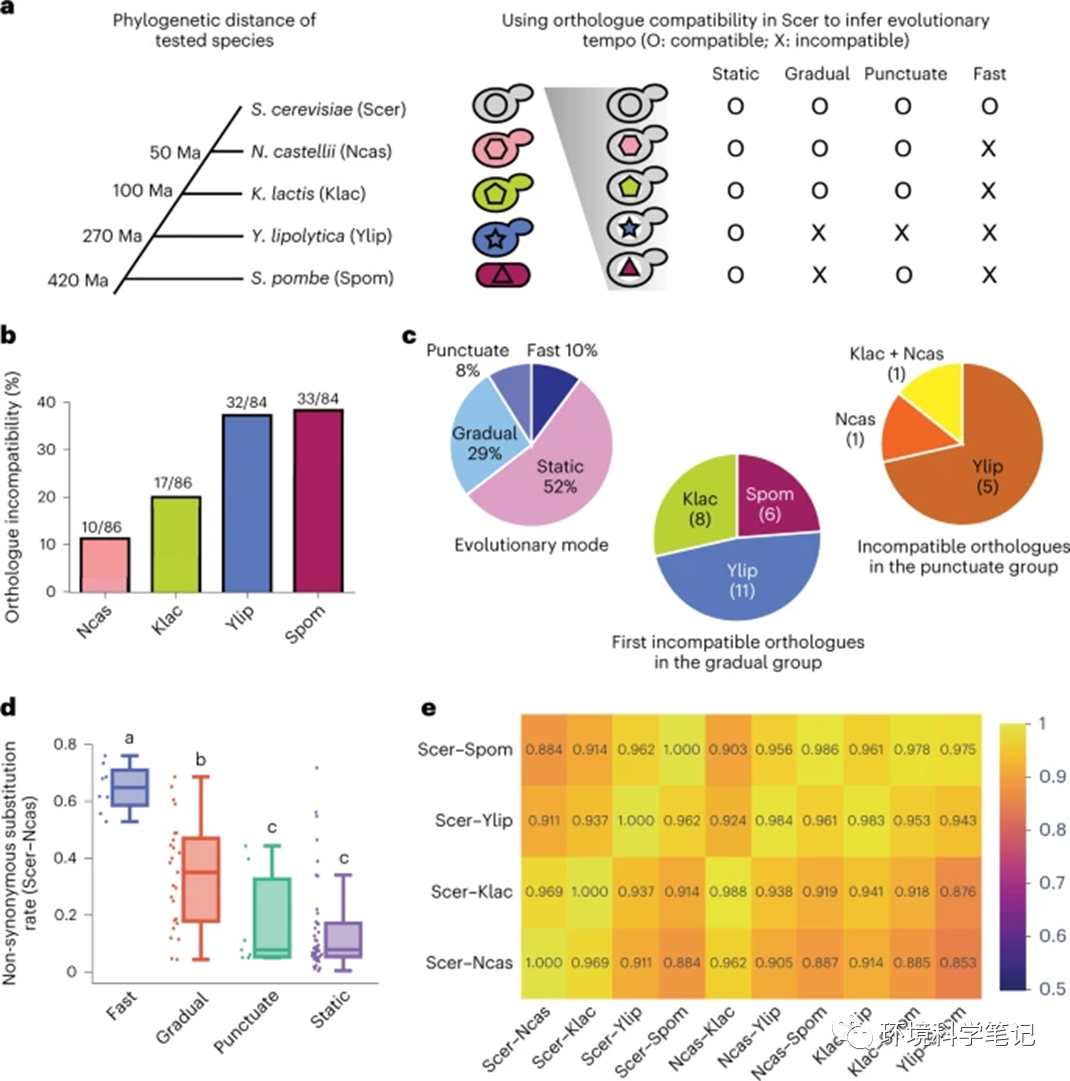
a, Orthologues from four additional Ascomycota species were used to study the evolution of essential genes. These four species diverged from S. cerevisiae over different timeframes, allowing us to characterize when the orthologues had become incompatible. To measure compatibility, S. cerevisiae essential genes were replaced by orthologues from the other four species and then cell viability was examined. Depending on the pattern of compatibility between orthologues, we classified the tested essential genes into four types: static, gradual, punctuate and fast. b, A total of 12 to 39% of tested orthologues are incompatible with the S. cerevisiae background. We performed a chi-square test to examine whether some species have more incompatible orthologues than the others and incompatible gene numbers are significantly different between different species (chi-square test, n = 340, P = 2.2 × 10−5). c, Distributions of essential genes exhibiting different evolutionary patterns. Only genes for which all four orthologues were tested are included in this analysis. Left: percentage of genes with different evolutionary patterns. Middle: the species when first incompatible orthologues were observed in the gradual group. Right: species-specific incompatibility in the punctuate group. Numbers of genes are shown in parentheses. d, The fast-type genes have the highest evolutionary rates between S. cerevisiae and N. castellii. Boxplots indicate median (middle line), 25th and 75th percentile (box), and minimum and maximum (whiskers). Distributions with different letters (above each boxplot) are significantly different from each other (fast: n = 8; gradual: n = 25; puncutate: n = 7; static: n = 44, two-sided Mann–Whitney U test, P values <0.05). See also the Source data for detailed statistical information. e, Most tested genes exhibit consistent evolutionary characteristics in all five species. Non-synonymous substitution rates (Ka) of tested orthologues from all species pairs were calculated (Supplementary Table 5) and high correlations of Ka between different species pairs were observed (one-tailed Spearman’s rank correlation, ρ = 0.85–0.99, P = 9.29 × 10−24–6.10 × 10−66; see Supplementary Table 6 for all P values). Spearman’s correlation coefficients are shown in the figure.
Fig. 2: Both protein-binding domains of Apc2 contribute to incompatibility.
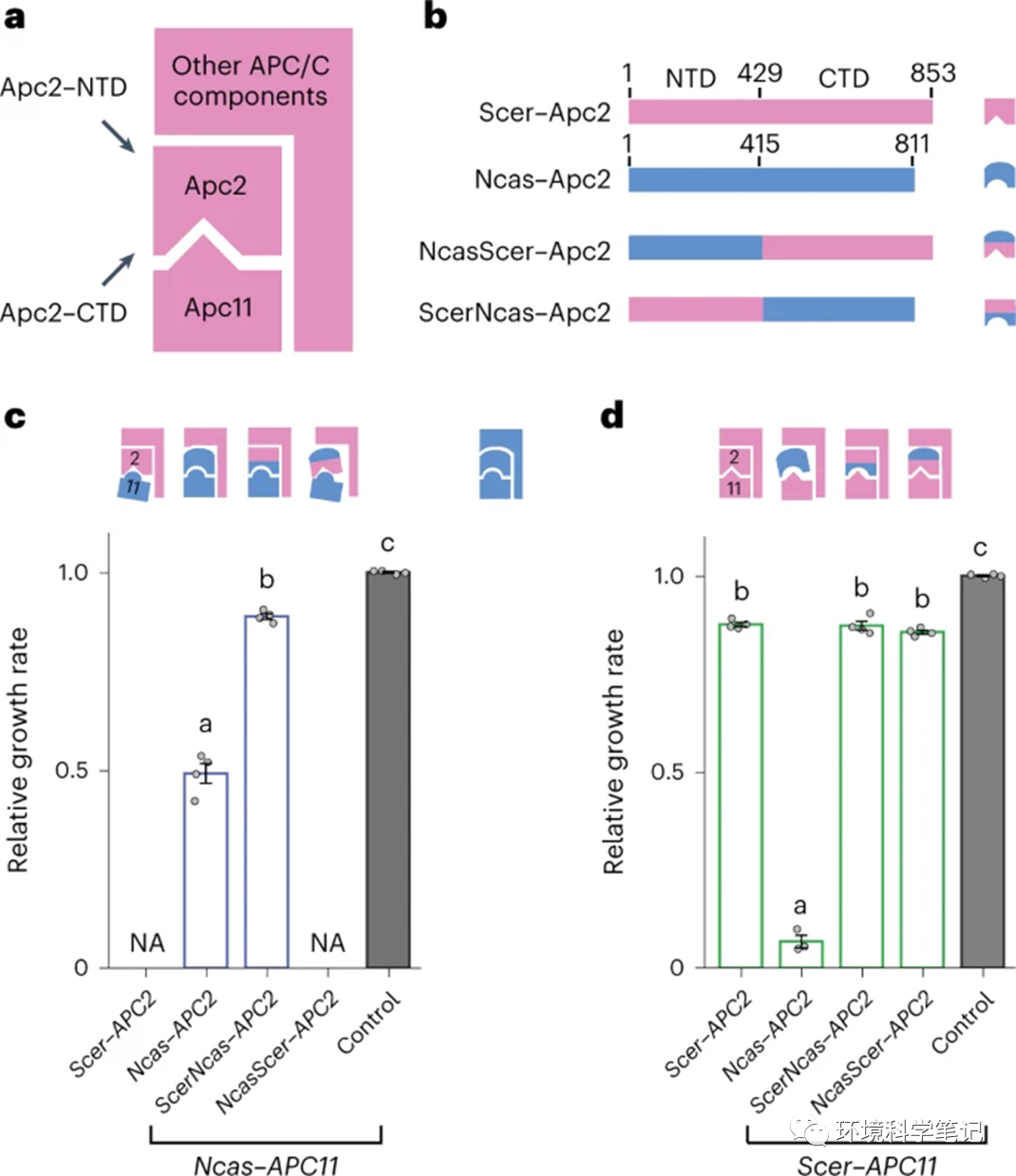
a, A simplified diagram depicting the interactions between Apc2, Apc11 and other APC/C components, which we employ in subsequent figures to illustrate how the structure of APC/C is affected by substituted orthologous proteins. The CTD of Apc2 is the main interacting domain with Apc11 and the NTD of Apc2 interacts with other APC/C components. b, Construction of chimeric proteins of Scer–Apc2 and Ncas–Apc2. N. castellii (Ncas) proteins are in blue and S. cerevisiae (Scer) proteins are in pink. c, Cells with the NTD of Scer–Apc2 or Ncas–Apc2 exhibit different fitness, suggesting that the NTD of Apc2 also contributes to the stability of the whole complex. The blue complex at top right represents the Ncas–APC/C. NA, data not available due to cell inviability. Control, the apc2Δ apc11Δ shuffling strain carrying the plasmid containing Scer–APC2 and Scer–APC11. d, Cells carrying ScerNcas–Apc2 and Scer–Apc11 do not have obvious growth defects, suggesting that the incompatibility of Ncas–Apc2 is not completely due to mis-interaction between the CTD of Ncas–Apc2 and Scer–Apc11. All compatibility tests of different proteins were performed in an apc2Δ apc11Δ shuffling strain hosting plasmids carrying the orthologous or chimeric genes being driven by the Tet-Off promoter. Growth rates were normalized to that of a control strain (apc2Δ apc11Δ + Scer–APC2 Scer–APC11). Error bars represent the standard error of the mean from four independent colonies. Columns sharing the same letters are not significantly different (ɑ = 0.05, Tukey-adjusted P values). Source data and detailed statistical information are provided in the Source data. See also Extended Data Fig. 5.
Fig. 3: A few diverged residues play a critical role in Apc11 orthologue incompatibility.
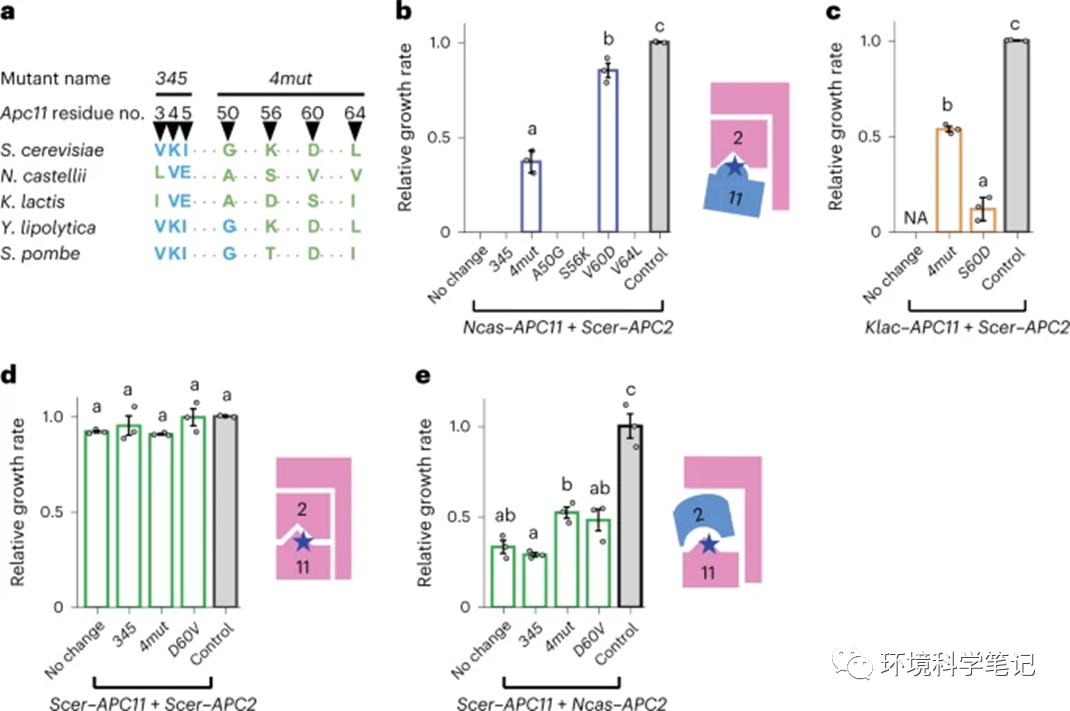
a, Seven residues in conserved domains of Apc11 were found to exhibit different patterns between compatible and incompatible orthologues. The amino acids shared by incompatible orthologues are in pink and those shared by compatible orthologues are in blue. Amino acids deviating from the majority of aligned orthologues are in green (Extended Data Fig. 7). b, Replacing residue 60 in Ncas–Apc11 rescues incompatibility with Scer–Apc2. Individual or multiple mutations were introduced into Ncas–APC11 to test for compatibility with Scer–Apc2. Only V60D and 4mut (quadruple substitution of residues 50, 56, 60 and 64) exhibited detectable rescue effects. Control, the apc2Δ apc11Δ shuffling strain carrying the plasmid containing Scer–APC2 and Scer–APC11. c, Replacing residue 60 in Klac–Apc11 only partially rescues incompatibility with Scer–Apc2, whereas Klac–APC11–4mut exhibits higher compatibility. d,e, Introduction of N. castellii residues into Scer–Apc11 does not result in obvious incompatibility with Scer–Apc2 (d). Nonetheless, these mutations increase the compatibility of Scer–Apc11 with Ncas–Apc2 (e), suggesting that residue 60 is crucial for the interaction between Apc11 and Apc2. All APC11 mutants and APC2 were driven by the Tet-Off promoter. Growth rates were normalized to that of the control strain. NA, data not available due to cell inviability. Error bars represent the standard error of the mean from three independent colonies. Columns sharing the same letters are not significantly different (ɑ = 0.05, Tukey-adjusted P values). Source data and detailed statistical information are provided in the Source data. Blue stars indicate the substituted residues in the interacting interface. No change means the template is the wild-type Ncas–Apc11 or Scer–Apc11.
Fig. 4: Effects ofAPC11 mutants depend on the stability of the protein complex.
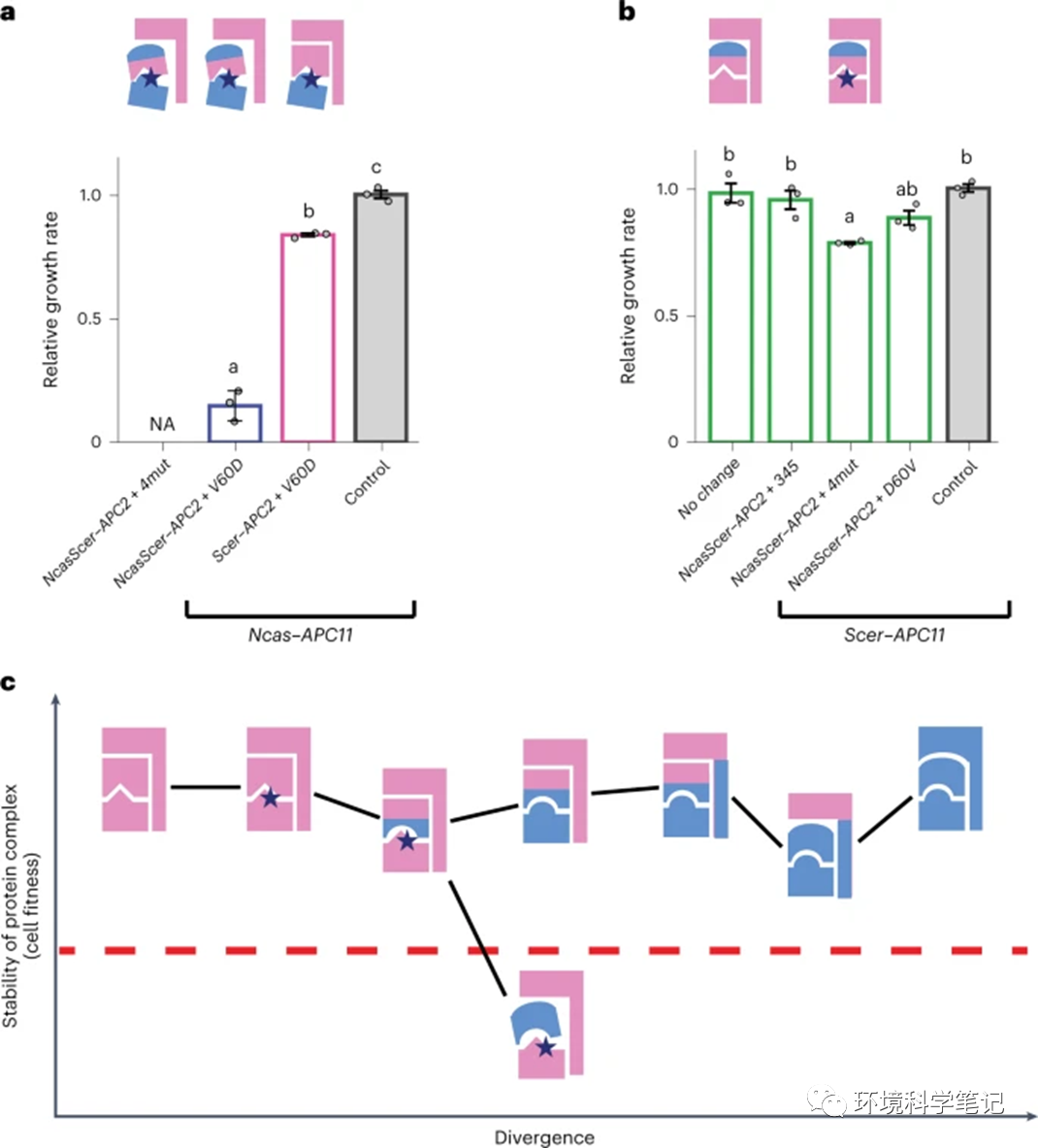
a, Ncas–APC11–V60D mutants exhibit different fitness in the NcasScer–APC2 and Scer–APC2 backgrounds. The Apc2–Apc11 interacting domains remain the same in these two strain backgrounds, but the APC/C complex is less stable in the NcasScer–APC2 background due to the foreign NTD of NcasScer–Apc2. b, Scer–APC11–4mut cells exhibit growth defects in the NcasScer–APC2 background. The 4mut and D60V mutants (see Fig. 3d) show no obvious effect in the Scer–APC2 background that has a more stable APC/C structure. Growth rates were normalized to that of the control strain. NA, data not available due to cell inviability. Error bars represent the standard error of the mean from three independent colonies. Columns sharing the same letters are not significantly different (ɑ = 0.05, Tukey-adjusted P values). Source data and detailed statistical information are provided in the Source data. Blue stars indicate the substituted residues in the interacting interface. No change means the template is the wild-type Ncas–Apc11 or Scer–Apc11. c, Illustration of the possible evolutionary trajectories of a protein complex under the influence of multiple protein interactions. Mutations accumulated in the interface of different subunits will be cryptically neutral if the structure of the complex is stably maintained by other components. As long as the effect of the mutations in the interface of primary or secondary interacting proteins does not exceed the buffering capacity of the complex (represented by the red dashed line), these mutations may accumulate in the population. Moreover, these mutations may allow interacting partners to further mutate, leading to co-evolution. However, phenotypic variation (or incompatibility) will be revealed if the stability of the complex is compromised sufficiently to exceed the buffering capacity.
Fig. 5: APC/C and its homologue, SCF, exhibit different evolutionary patterns.
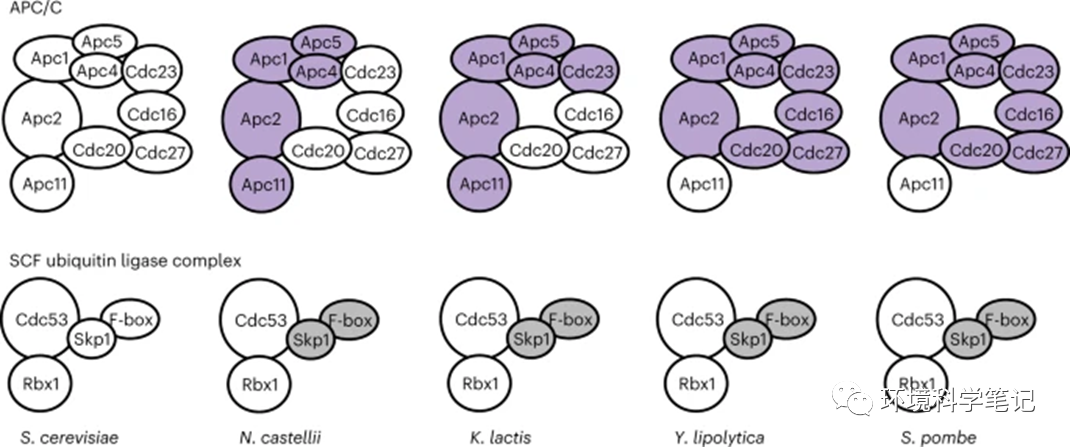
Both APC/C and SCF complexes are the ubiquitin E3 ligases involved in cell cycle progression, but APC/C has far more subunits than SFC. Most APC/C core subunits evolve incompatibility betweenS. cerevisiae and N. castellii. In contrast, SCF core subunits remain static in all species tested (Supplementary Table 1). Orthologous APC/C subunits incompatible with S. cerevisiae are coloured purple and uncharacterized SCF subunits are coloured grey.
